Note
Go to the end to download the full example code
Example: Deep Markov Model inferred using SteinVI¶
In this example we infer a deep Markov model (DMM) using SteinVI for generating music (chorales by Johan Sebastian Bach).
The model DMM based on reference [1][2] and the Pyro DMM example: https://pyro.ai/examples/dmm.html.
Reference:
Pathwise Derivatives for Multivariate Distributions Martin Jankowiak and Theofanis Karaletsos (2019)
- Structured Inference Networks for Nonlinear State Space Models [arXiv:1609.09869]
Rahul G. Krishnan, Uri Shalit and David Sontag (2016)
import argparse
import numpy as np
import jax
from jax import nn, numpy as jnp, random
from optax import adam, chain
import numpyro
from numpyro.contrib.einstein import SteinVI
from numpyro.contrib.einstein.mixture_guide_predictive import MixtureGuidePredictive
from numpyro.contrib.einstein.stein_kernels import RBFKernel
import numpyro.distributions as dist
from numpyro.examples.datasets import JSB_CHORALES, load_dataset
from numpyro.optim import optax_to_numpyro
def _reverse_padded(padded, lengths):
def _reverse_single(p, length):
new = jnp.zeros_like(p)
reverse = jnp.roll(p[::-1], length, axis=0)
return new.at[:].set(reverse)
return jax.vmap(_reverse_single)(padded, lengths)
def load_data(split="train"):
_, fetch = load_dataset(JSB_CHORALES, split=split)
lengths, seqs = fetch(0)
return (seqs, _reverse_padded(seqs, lengths), lengths)
def emitter(x, params):
"""Parameterizes the bernoulli observation likelihood `p(x_t | z_t)`"""
l1 = nn.relu(jnp.matmul(x, params["l1"]))
l2 = nn.relu(jnp.matmul(l1, params["l2"]))
return jnp.matmul(l2, params["l3"])
def transition(x, params):
"""Parameterizes the gaussian latent transition probability `p(z_t | z_{t-1})`
See section 5 in [1].
**Reference:**
1. Structured Inference Networks for Nonlinear State Space Models [arXiv:1609.09869]
Rahul G. Krishnan, Uri Shalit and David Sontag (2016)
"""
def _gate(x, params):
l1 = nn.relu(jnp.matmul(x, params["l1"]))
return nn.sigmoid(jnp.matmul(l1, params["l2"]))
def _shared(x, params):
l1 = nn.relu(jnp.matmul(x, params["l1"]))
return jnp.matmul(l1, params["l2"])
def _mean(x, params):
return jnp.matmul(x, params["l1"])
def _std(x, params):
l1 = jnp.matmul(nn.relu(x), params["l1"])
return nn.softplus(l1)
gt = _gate(x, params["gate"])
ht = _shared(x, params["shared"])
loc = (1 - gt) * _mean(x, params["mean"]) + gt * ht
std = _std(ht, params["std"])
return loc, std
def combiner(x, params):
mean = jnp.matmul(x, params["mean"])
std = nn.softplus(jnp.matmul(x, params["std"]))
return mean, std
def gru(xs, lengths, init_hidden, params):
"""RNN with GRU. Based on https://github.com/google/jax/pull/2298"""
def apply_fun_single(state, inputs):
i, x = inputs
inp_update = jnp.matmul(x, params["update_in"])
hidden_update = jnp.dot(state, params["update_weight"])
update_gate = nn.sigmoid(inp_update + hidden_update)
reset_gate = nn.sigmoid(
jnp.matmul(x, params["reset_in"]) + jnp.dot(state, params["reset_weight"])
)
output_gate = update_gate * state + (1 - update_gate) * jnp.tanh(
jnp.matmul(x, params["out_in"])
+ jnp.dot(reset_gate * state, params["out_weight"])
)
hidden = jnp.where((i < lengths)[:, None], output_gate, jnp.zeros_like(state))
return hidden, hidden
init_hidden = jnp.broadcast_to(init_hidden, (xs.shape[1], init_hidden.shape[1]))
return jax.lax.scan(apply_fun_single, init_hidden, (jnp.arange(xs.shape[0]), xs))
def _normal_init(*shape):
return lambda rng_key: dist.Normal(scale=0.1).sample(rng_key, shape)
def model(
seqs,
seqs_rev,
lengths,
*,
subsample_size=77,
latent_dim=32,
emission_dim=100,
transition_dim=200,
data_dim=88,
gru_dim=150,
annealing_factor=1.0,
predict=False,
):
max_seq_length = seqs.shape[1]
emitter_params = {
"l1": numpyro.param("emitter_l1", _normal_init(latent_dim, emission_dim)),
"l2": numpyro.param("emitter_l2", _normal_init(emission_dim, emission_dim)),
"l3": numpyro.param("emitter_l3", _normal_init(emission_dim, data_dim)),
}
trans_params = {
"gate": {
"l1": numpyro.param("gate_l1", _normal_init(latent_dim, transition_dim)),
"l2": numpyro.param("gate_l2", _normal_init(transition_dim, latent_dim)),
},
"shared": {
"l1": numpyro.param("shared_l1", _normal_init(latent_dim, transition_dim)),
"l2": numpyro.param("shared_l2", _normal_init(transition_dim, latent_dim)),
},
"mean": {"l1": numpyro.param("mean_l1", _normal_init(latent_dim, latent_dim))},
"std": {"l1": numpyro.param("std_l1", _normal_init(latent_dim, latent_dim))},
}
z0 = numpyro.param(
"z0", lambda rng_key: dist.Normal(0, 1.0).sample(rng_key, (latent_dim,))
)
z0 = jnp.broadcast_to(z0, (subsample_size, 1, latent_dim))
with numpyro.plate(
"data", seqs.shape[0], subsample_size=subsample_size, dim=-1
) as idx:
if subsample_size == seqs.shape[0]:
seqs_batch = seqs
lengths_batch = lengths
else:
seqs_batch = seqs[idx]
lengths_batch = lengths[idx]
masks = jnp.repeat(
jnp.expand_dims(jnp.arange(max_seq_length), axis=0), subsample_size, axis=0
) < jnp.expand_dims(lengths_batch, axis=-1)
# NB: Mask is to avoid scoring 'z' using distribution at this point
z = numpyro.sample(
"z",
dist.Normal(0.0, jnp.ones((max_seq_length, latent_dim)))
.mask(False)
.to_event(2),
)
z_shift = jnp.concatenate([z0, z[:, :-1, :]], axis=-2)
z_loc, z_scale = transition(z_shift, params=trans_params)
with numpyro.handlers.scale(scale=annealing_factor):
# Actually score 'z'
numpyro.sample(
"z_aux",
dist.Normal(z_loc, z_scale)
.mask(jnp.expand_dims(masks, axis=-1))
.to_event(2),
obs=z,
)
emission_probs = emitter(z, params=emitter_params)
if predict:
tunes = None
else:
tunes = seqs_batch
numpyro.sample(
"tunes",
dist.Bernoulli(logits=emission_probs)
.mask(jnp.expand_dims(masks, axis=-1))
.to_event(2),
obs=tunes,
)
def guide(
seqs,
seqs_rev,
lengths,
*,
subsample_size=77,
latent_dim=32,
emission_dim=100,
transition_dim=200,
data_dim=88,
gru_dim=150,
annealing_factor=1.0,
predict=False,
):
max_seq_length = seqs.shape[1]
seqs_rev = jnp.transpose(seqs_rev, axes=(1, 0, 2))
combiner_params = {
"mean": numpyro.param("combiner_mean", _normal_init(gru_dim, latent_dim)),
"std": numpyro.param("combiner_std", _normal_init(gru_dim, latent_dim)),
}
gru_params = {
"update_in": numpyro.param("update_in", _normal_init(data_dim, gru_dim)),
"update_weight": numpyro.param("update_weight", _normal_init(gru_dim, gru_dim)),
"reset_in": numpyro.param("reset_in", _normal_init(data_dim, gru_dim)),
"reset_weight": numpyro.param("reset_weight", _normal_init(gru_dim, gru_dim)),
"out_in": numpyro.param("out_in", _normal_init(data_dim, gru_dim)),
"out_weight": numpyro.param("out_weight", _normal_init(gru_dim, gru_dim)),
}
with numpyro.plate(
"data", seqs.shape[0], subsample_size=subsample_size, dim=-1
) as idx:
if subsample_size == seqs.shape[0]:
seqs_rev_batch = seqs_rev
lengths_batch = lengths
else:
seqs_rev_batch = seqs_rev[:, idx, :]
lengths_batch = lengths[idx]
masks = jnp.repeat(
jnp.expand_dims(jnp.arange(max_seq_length), axis=0), subsample_size, axis=0
) < jnp.expand_dims(lengths_batch, axis=-1)
h0 = numpyro.param(
"h0",
lambda rng_key: dist.Normal(0.0, 1).sample(rng_key, (1, gru_dim)),
)
_, hs = gru(seqs_rev_batch, lengths_batch, h0, gru_params)
hs = _reverse_padded(jnp.transpose(hs, axes=(1, 0, 2)), lengths_batch)
with numpyro.handlers.scale(scale=annealing_factor):
numpyro.sample(
"z",
dist.Normal(*combiner(hs, combiner_params))
.mask(jnp.expand_dims(masks, axis=-1))
.to_event(2),
)
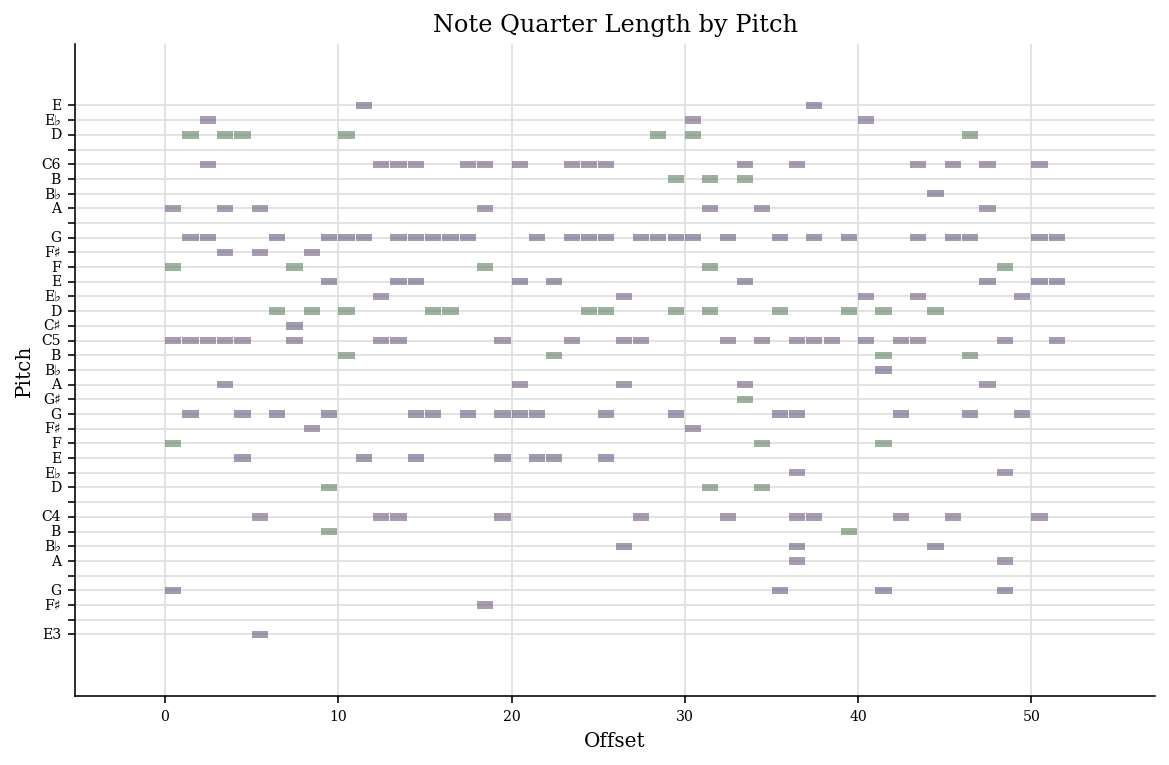
def vis_tune(i, tunes, lengths, name="stein_dmm.pdf"):
tune = tunes[i, : lengths[i]]
try:
from music21.chord import Chord
from music21.pitch import Pitch
from music21.stream import Stream
stream = Stream()
for chord in tune:
stream.append(
Chord(list(Pitch(pitch) for pitch in (np.arange(88) + 21)[chord > 0]))
)
plot = stream.plot(doneAction=None)
plot.write(name)
except ModuleNotFoundError:
import matplotlib.pyplot as plt
plt.imshow(tune.T, cmap="Greys")
plt.ylabel("Pitch")
plt.xlabel("Offset")
plt.savefig(name)
def main(args):
inf_key, pred_key = random.split(random.PRNGKey(seed=args.rng_seed), 2)
steinvi = SteinVI(
model,
guide,
optax_to_numpyro(chain(adam(1e-2))),
RBFKernel(),
num_elbo_particles=args.num_elbo_particles,
num_stein_particles=args.num_stein_particles,
)
seqs, rev_seqs, lengths = load_data()
results = steinvi.run(
inf_key,
args.max_iter,
seqs,
rev_seqs,
lengths,
gru_dim=args.gru_dim,
subsample_size=args.subsample_size,
)
pred = MixtureGuidePredictive(
model,
guide,
params=results.params,
num_samples=1,
guide_sites=steinvi.guide_sites,
)
seqs, rev_seqs, lengths = load_data("valid")
pred_notes = pred(
pred_key, seqs, rev_seqs, lengths, subsample_size=seqs.shape[0], predict=True
)["tunes"]
vis_tune(0, pred_notes[0], lengths)
if __name__ == "__main__":
parser = argparse.ArgumentParser()
parser.add_argument("--subsample-size", type=int, default=10)
parser.add_argument("--max-iter", type=int, default=100)
parser.add_argument("--repulsion", type=float, default=1.0)
parser.add_argument("--verbose", type=bool, default=True)
parser.add_argument("--num-stein-particles", type=int, default=5)
parser.add_argument("--num-elbo-particles", type=int, default=5)
parser.add_argument("--progress-bar", type=bool, default=True)
parser.add_argument("--gru-dim", type=int, default=150)
parser.add_argument("--rng-key", type=int, default=142)
parser.add_argument("--device", default="cpu", choices=["gpu", "cpu"])
parser.add_argument("--rng-seed", default=142, type=int)
args = parser.parse_args()
numpyro.set_platform(args.device)
main(args)