Note
Go to the end to download the full example code
Example: Hamiltonian Monte Carlo with Energy Conserving Subsampling¶
This example illustrates the use of data subsampling in HMC using Energy Conserving Subsampling. Data subsampling is applicable when the likelihood factorizes as a product of N terms.
References:
Hamiltonian Monte Carlo with energy conserving subsampling, Dang, K. D., Quiroz, M., Kohn, R., Minh-Ngoc, T., & Villani, M. (2019)
import argparse
import time
import matplotlib.pyplot as plt
import numpy as np
from jax import random
import jax.numpy as jnp
import numpyro
import numpyro.distributions as dist
from numpyro.examples.datasets import HIGGS, load_dataset
from numpyro.infer import HMC, HMCECS, MCMC, NUTS, SVI, Trace_ELBO, autoguide
def model(data, obs, subsample_size):
n, m = data.shape
theta = numpyro.sample("theta", dist.Normal(jnp.zeros(m), 0.5 * jnp.ones(m)))
with numpyro.plate("N", n, subsample_size=subsample_size):
batch_feats = numpyro.subsample(data, event_dim=1)
batch_obs = numpyro.subsample(obs, event_dim=0)
numpyro.sample(
"obs", dist.Bernoulli(logits=theta @ batch_feats.T), obs=batch_obs
)
def run_hmcecs(hmcecs_key, args, data, obs, inner_kernel):
svi_key, mcmc_key = random.split(hmcecs_key)
# find reference parameters for second order taylor expansion to estimate likelihood (taylor_proxy)
optimizer = numpyro.optim.Adam(step_size=1e-3)
guide = autoguide.AutoDelta(model)
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
svi_result = svi.run(svi_key, args.num_svi_steps, data, obs, args.subsample_size)
params, losses = svi_result.params, svi_result.losses
ref_params = {"theta": params["theta_auto_loc"]}
# taylor proxy estimates log likelihood (ll) by
# taylor_expansion(ll, theta_curr) +
# sum_{i in subsample} ll_i(theta_curr) - taylor_expansion(ll_i, theta_curr) around ref_params
proxy = HMCECS.taylor_proxy(ref_params)
kernel = HMCECS(inner_kernel, num_blocks=args.num_blocks, proxy=proxy)
mcmc = MCMC(kernel, num_warmup=args.num_warmup, num_samples=args.num_samples)
mcmc.run(mcmc_key, data, obs, args.subsample_size)
mcmc.print_summary()
return losses, mcmc.get_samples()
def run_hmc(mcmc_key, args, data, obs, kernel):
mcmc = MCMC(kernel, num_warmup=args.num_warmup, num_samples=args.num_samples)
mcmc.run(mcmc_key, data, obs, None)
mcmc.print_summary()
return mcmc.get_samples()
def main(args):
assert (
11_000_000 >= args.num_datapoints
), "11,000,000 data points in the Higgs dataset"
# full dataset takes hours for plain hmc!
if args.dataset == "higgs":
_, fetch = load_dataset(
HIGGS, shuffle=False, num_datapoints=args.num_datapoints
)
data, obs = fetch()
else:
data, obs = (np.random.normal(size=(10, 28)), np.ones(10))
hmcecs_key, hmc_key = random.split(random.PRNGKey(args.rng_seed))
# choose inner_kernel
if args.inner_kernel == "hmc":
inner_kernel = HMC(model)
else:
inner_kernel = NUTS(model)
start = time.time()
losses, hmcecs_samples = run_hmcecs(hmcecs_key, args, data, obs, inner_kernel)
hmcecs_runtime = time.time() - start
start = time.time()
hmc_samples = run_hmc(hmc_key, args, data, obs, inner_kernel)
hmc_runtime = time.time() - start
summary_plot(losses, hmc_samples, hmcecs_samples, hmc_runtime, hmcecs_runtime)
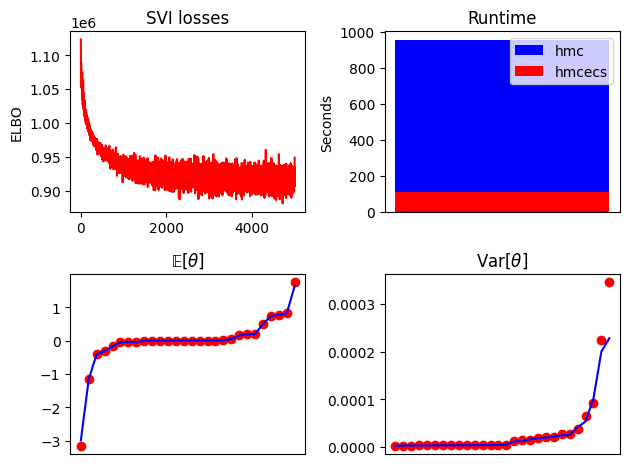
def summary_plot(losses, hmc_samples, hmcecs_samples, hmc_runtime, hmcecs_runtime):
fig, ax = plt.subplots(2, 2)
ax[0, 0].plot(losses, "r")
ax[0, 0].set_title("SVI losses")
ax[0, 0].set_ylabel("ELBO")
if hmc_runtime > hmcecs_runtime:
ax[0, 1].bar([0], hmc_runtime, label="hmc", color="b")
ax[0, 1].bar([0], hmcecs_runtime, label="hmcecs", color="r")
else:
ax[0, 1].bar([0], hmcecs_runtime, label="hmcecs", color="r")
ax[0, 1].bar([0], hmc_runtime, label="hmc", color="b")
ax[0, 1].set_title("Runtime")
ax[0, 1].set_ylabel("Seconds")
ax[0, 1].legend()
ax[0, 1].set_xticks([])
ax[1, 0].plot(jnp.sort(hmc_samples["theta"].mean(0)), "or")
ax[1, 0].plot(jnp.sort(hmcecs_samples["theta"].mean(0)), "b")
ax[1, 0].set_title(r"$\mathrm{\mathbb{E}}[\theta]$")
ax[1, 1].plot(jnp.sort(hmc_samples["theta"].var(0)), "or")
ax[1, 1].plot(jnp.sort(hmcecs_samples["theta"].var(0)), "b")
ax[1, 1].set_title(r"Var$[\theta]$")
for a in ax[1, :]:
a.set_xticks([])
fig.tight_layout()
fig.savefig("hmcecs_plot.pdf", bbox_inches="tight")
if __name__ == "__main__":
parser = argparse.ArgumentParser(
"Hamiltonian Monte Carlo with Energy Conserving Subsampling"
)
parser.add_argument("--subsample_size", type=int, default=1300)
parser.add_argument("--num_svi_steps", type=int, default=5000)
parser.add_argument("--num_blocks", type=int, default=100)
parser.add_argument("--num_warmup", type=int, default=500)
parser.add_argument("--num_samples", type=int, default=500)
parser.add_argument("--num_datapoints", type=int, default=1_500_000)
parser.add_argument(
"--dataset", type=str, choices=["higgs", "mock"], default="higgs"
)
parser.add_argument(
"--inner_kernel", type=str, choices=["nuts", "hmc"], default="nuts"
)
parser.add_argument("--device", default="cpu", type=str, choices=["cpu", "gpu"])
parser.add_argument(
"--rng_seed", default=37, type=int, help="random number generator seed"
)
args = parser.parse_args()
numpyro.set_platform(args.device)
main(args)