Note
Click here to download the full example code
Example: Neal’s Funnel¶
This example, which is adapted from [1], illustrates how to leverage non-centered
parameterization using the reparam handler.
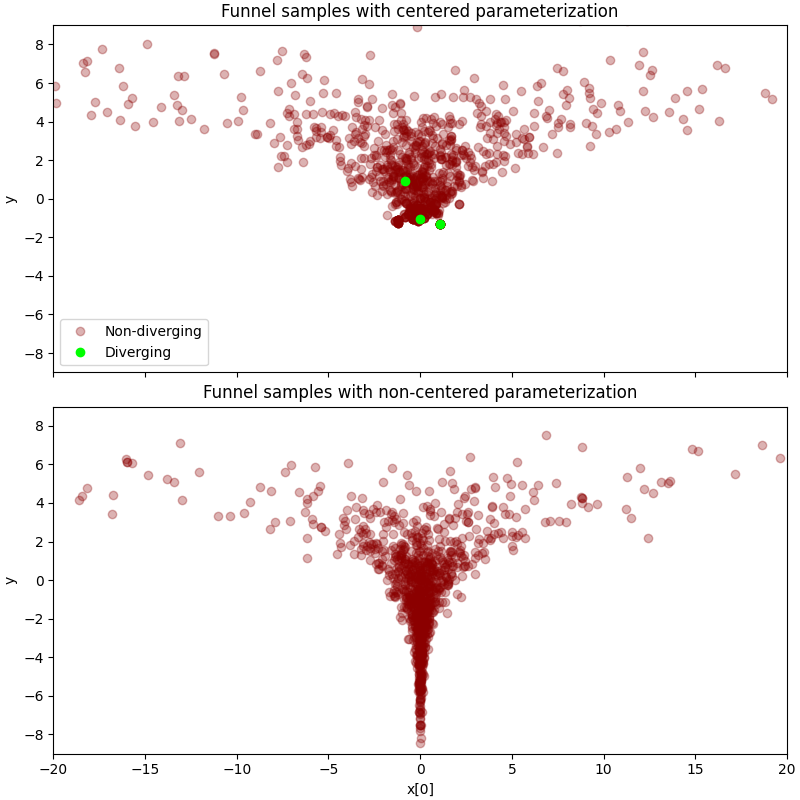
We will examine the difference between two types of parameterizations on the
10-dimensional Neal’s funnel distribution. As we will see, HMC gets trouble at
the neck of the funnel if centered parameterization is used. On the contrary,
the problem can be solved by using non-centered parameterization.
Using non-centered parameterization through LocScaleReparam
or TransformReparam in NumPyro has the same effect as
the automatic reparameterisation technique introduced in [2].
References:
- Stan User’s Guide, https://mc-stan.org/docs/2_19/stan-users-guide/reparameterization-section.html
- Maria I. Gorinova, Dave Moore, Matthew D. Hoffman (2019), “Automatic Reparameterisation of Probabilistic Programs”, (https://arxiv.org/abs/1906.03028)
import argparse
import os
import matplotlib.pyplot as plt
from jax import random
import jax.numpy as jnp
import numpyro
import numpyro.distributions as dist
from numpyro.handlers import reparam
from numpyro.infer import MCMC, NUTS, Predictive
from numpyro.infer.reparam import LocScaleReparam
def model(dim=10):
y = numpyro.sample("y", dist.Normal(0, 3))
numpyro.sample("x", dist.Normal(jnp.zeros(dim - 1), jnp.exp(y / 2)))
reparam_model = reparam(model, config={"x": LocScaleReparam(0)})
def run_inference(model, args, rng_key):
kernel = NUTS(model)
mcmc = MCMC(
kernel,
num_warmup=args.num_warmup,
num_samples=args.num_samples,
num_chains=args.num_chains,
progress_bar=False if "NUMPYRO_SPHINXBUILD" in os.environ else True,
)
mcmc.run(rng_key)
mcmc.print_summary(exclude_deterministic=False)
return mcmc
def main(args):
rng_key = random.PRNGKey(0)
# do inference with centered parameterization
print(
"============================= Centered Parameterization =============================="
)
mcmc = run_inference(model, args, rng_key)
samples = mcmc.get_samples()
diverging = mcmc.get_extra_fields()["diverging"]
# do inference with non-centered parameterization
print(
"\n=========================== Non-centered Parameterization ============================"
)
reparam_mcmc = run_inference(reparam_model, args, rng_key)
reparam_samples = reparam_mcmc.get_samples()
reparam_diverging = reparam_mcmc.get_extra_fields()["diverging"]
# collect deterministic sites
reparam_samples = Predictive(
reparam_model, reparam_samples, return_sites=["x", "y"]
)(random.PRNGKey(1))
# make plots
fig, (ax1, ax2) = plt.subplots(
2, 1, sharex=True, figsize=(8, 8), constrained_layout=True
)
ax1.plot(
samples["x"][~diverging, 0],
samples["y"][~diverging],
"o",
color="darkred",
alpha=0.3,
label="Non-diverging",
)
ax1.plot(
samples["x"][diverging, 0],
samples["y"][diverging],
"o",
color="lime",
label="Diverging",
)
ax1.set(
xlim=(-20, 20),
ylim=(-9, 9),
ylabel="y",
title="Funnel samples with centered parameterization",
)
ax1.legend()
ax2.plot(
reparam_samples["x"][~reparam_diverging, 0],
reparam_samples["y"][~reparam_diverging],
"o",
color="darkred",
alpha=0.3,
)
ax2.plot(
reparam_samples["x"][reparam_diverging, 0],
reparam_samples["y"][reparam_diverging],
"o",
color="lime",
)
ax2.set(
xlim=(-20, 20),
ylim=(-9, 9),
xlabel="x[0]",
ylabel="y",
title="Funnel samples with non-centered parameterization",
)
plt.savefig("funnel_plot.pdf")
if __name__ == "__main__":
assert numpyro.__version__.startswith("0.12.0")
parser = argparse.ArgumentParser(
description="Non-centered reparameterization example"
)
parser.add_argument("-n", "--num-samples", nargs="?", default=1000, type=int)
parser.add_argument("--num-warmup", nargs="?", default=1000, type=int)
parser.add_argument("--num-chains", nargs="?", default=1, type=int)
parser.add_argument("--device", default="cpu", type=str, help='use "cpu" or "gpu".')
args = parser.parse_args()
numpyro.set_platform(args.device)
numpyro.set_host_device_count(args.num_chains)
main(args)